SVMerge
We present a pipeline, SVMerge, to detect structural variants (SVs) by integrating calls from several existing SV callers, which are then validated and the breakpoints refined using local de novo assembly. SVMerge is modular and extensible allowing new callers to be incorporated as they become available.
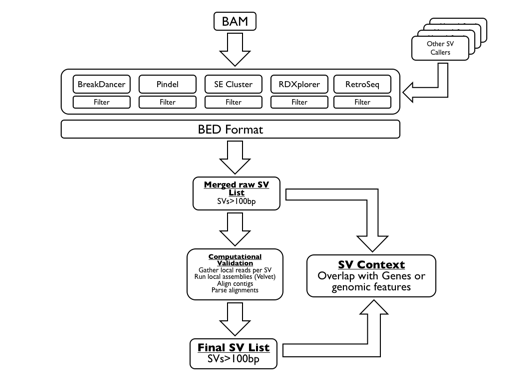 SVMerge uses a suite of software tools to detect structural variants (SVs) from mapped reads. The calls are filtered, merged and then validated computationally by local de novo assembly. The output is in BED format, allowing for easy downstream analysis or viewing in a genome browser. The SVMerge pipeline is extendable so that calls made by other packages can be included in the downstream analysis.
SVMerge uses a suite of software tools to detect structural variants (SVs) from mapped reads. The calls are filtered, merged and then validated computationally by local de novo assembly. The output is in BED format, allowing for easy downstream analysis or viewing in a genome browser. The SVMerge pipeline is extendable so that calls made by other packages can be included in the downstream analysis.
Download
The source code and binaries for SVMerge are available to download from the sourceforge download page.
Documentation
The user manual for SVMerge is available for download from here.
Reference
To cite SVMerge:
Wong K, Keane TM, Stalker J, Adams DJ (2010) SVMerge: Enhanced structural variant and breakpoint detection by integration of multiple detection methods and local assembly, Genome Biology, 11:R128 link
The SV calls for the human trio that was analysed in the paper are available for download from here
Help
If you have any problems running SVMerge, please contact us by leaving a message on our support forum or you can email the main developer, Kim Wong (kw10 at sanger dot ac dot uk).